
SDAP Home Page
SDAP Overview
SDAP All
SDAP Food
Use SDAP All
Search SDAP All
List SDAP All
SDAP Food
SDAP Tools
FAO/WHO Allergenicity Test
FASTA Search in SDAP
Peptide Match
Peptide Similarity
Peptide-Protein PD Index
Aller_ML, Allergen Markup Language
List SDAP
About SDAP
General Information
Manual
FAQ
Publications
Who Are We
Advisory Board
Allergy Links
Our Software Tools
MPACK
FANTOM
GETAREA
NOAH/DIAMOD
MASIA
PCPMer
InterProSurf
EpiSearch
Protein Databases
PDB
MMDB - Entrez
SWISS-PROT
NCBI - Entrez
PIR
Protein Classification
CATH
CE
FSSP
iProClass
ProtoMap
SCOP
TOPS
VAST
Bioinformatics Servers
@TOME
BLAST @ NCBI
BLAST @ PIR
FASTA @ PIR
Peptide Match @ PIR
ClustalW @ BCM
ClustalW @ EMBL - EBI
ClustalW @ PIR
Bioinformatics Tools
Cn3D
MolMol
Bioinformatics Links
Bioinformatics Links Directory
|
SDAP - All Allergens
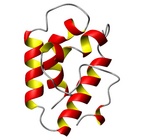
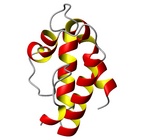 | Allergen Par j 2Translate to AllerML | 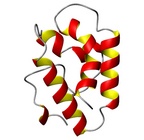 | 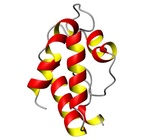 | | Allergen | Par j 2 | | Type | weed | | Species - Systematic Name | Rosales; Parietaria judaica | | Species - Common Name | Pellitory-of-the-Wall | | Keywords | lipid transfer protein; P2 protein
| | Class | IUIS |
|  |
Par j 2 - Isoallergens
Par j 2 - Protein Sequences
FASTA@SDAP: FASTA search against all SDAP allergen sequences performed at SDAP
BLAST@ExPASy: BLAST search performed at the Expert Protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (SIB)
PROSITE@PIR: PROSITE search performed at PIR - Protein Information Resources
Par j 2 - Protein Sequence Properties
PeptideCutter@ExPASy: Protease cleavage sites predicted with PeptideCutter from the Expert Protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (SIB)
Par j 2 - Protein Similarity, Classification and Clustering
Par j 2 - Protein Models
Par j 2 - Pfam domains
| Sequence | Pfam sequence | Pfam family | Allergens from
this Pfam family | Pfam database | Pfam region | First amino acid | Last amino acid | | P55958 | | | | | | | | | | NL21_PARJU | PF00234 | Go! | Pfam A | 1 | 35 | 121 | | | | PF00234: Protease inhibitor/seed storage/LTP family
| | O04403 | | | | | | | | | | NL22_PARJU | PF00234 | Go! | Pfam A | 1 | 35 | 121 | | | | PF00234: Protease inhibitor/seed storage/LTP family
|
Pfam is a database of multiple sequence alignments and hidden Markov models covering protein domains and families
Inspect MotifMate results for Par j 2. AutoMotif contains a database of PCPMer motifs for allergens.
MotifMate home page
MotifMate summary
Browse MotifMate proteins
Browse MotifMate families
Par j 2 - EpitopesJ. A. Asturias, N. Gomez-Bayon, J. L. Eseverri, and A. Martinez,
Par j 1 and Par j 2, the major allergens from Parietaria judaica pollen,
have similar immunoglobulin E epitopes,
Clin. Exp. Allergy 2003, 33, 518-524.
|

