
SDAP Home Page
SDAP Overview
Search SDAP
SDAP All
SDAP Food
SDAP Tools
AllergenAI
FAO/WHO Allergenicity Test
FASTA Search in SDAP
Peptide Match
Peptide Similarity
Peptide-Protein PD Index
Aller_ML, Allergen Markup Language
List SDAP
About SDAP
General Information
Manual
FAQ
Publications
Who Are We
Advisory Board
New Allergen Submission form
Allergy Links
Our Software Tools
MPACK
FANTOM
GETAREA
InterProSurf
EpiSearch
Allergen Databases
WHO/IUIS Allergen Nomenclature database
FARRP Allergen Protein Database (University of Nebraska)
Allergen Database for Food Safety (ADFS)
COMPARE database
ALLFAM (Medical University of Vienna)
Allermatch (Wageninen University)
Protein Databases
PDB
MMDB - Entrez
SWISS-PROT
NCBI - Entrez
PIR
Protein Classification
CATH
FSSP
iProClass
ProtoMap
SCOP
VAST
Bioinformatics Servers
BLAST @ NCBI
FASTA @ EMBL-EBI
Peptide Match @ PIR
ClustalW @ EMBL - EBI
Bioinformatics Tools
Cn3D
|
SDAP 2.0 - Structural Database of Allergenic Proteins
E1-E5 Property-Based Peptide Similarity Index PD for Two Sequences
Compute the PD sequence similarity index for two sequences:
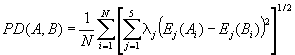
where λj is the eigenvalue of the j-th E
component, Ej(Ai) is the Ej
value for the amino acid in the i-th position from sequence A,
and Ej(Bi) is the Ej
value for the amino acid in the i-th position from sequence B.
The first sequence should be shorter
or equal to the second sequence. If the first sequence is shorter,
then the first sequence is moved along the second sequence, one position at a time, and all PD
values are reported.
Enter each protein sequence as a string of single-letter amino acid codes
(the maximum length of the sequence is 1000). Sequences longer than 1000 amino acids will
be truncated after the first 1000 amino acids. Gaps must be indicated by dashes (-).
The symbols B, U, Z, X, *, and . are automatically converted to - (gap).
Any other symbol will be deleted from the sequence, while the presence of other letters
will trigger the stop of the computation.
|

