
SDAP Home Page
SDAP Overview
SDAP All
SDAP Food
Use SDAP All
Search SDAP All
List SDAP All
SDAP Food
SDAP Tools
FAO/WHO Allergenicity Test
FASTA Search in SDAP
Peptide Match
Peptide Similarity
Peptide-Protein PD Index
Aller_ML, Allergen Markup Language
List SDAP
About SDAP
General Information
Manual
FAQ
Publications
Who Are We
Advisory Board
Allergy Links
Our Software Tools
MPACK
FANTOM
GETAREA
NOAH/DIAMOD
MASIA
PCPMer
InterProSurf
EpiSearch
Protein Databases
PDB
MMDB - Entrez
SWISS-PROT
NCBI - Entrez
PIR
Protein Classification
CATH
CE
FSSP
iProClass
ProtoMap
SCOP
TOPS
VAST
Bioinformatics Servers
@TOME
BLAST @ NCBI
BLAST @ PIR
FASTA @ PIR
Peptide Match @ PIR
ClustalW @ BCM
ClustalW @ EMBL - EBI
ClustalW @ PIR
Bioinformatics Tools
Cn3D
MolMol
Bioinformatics Links
Bioinformatics Links Directory
|
SDAP - All Allergens
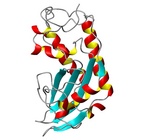 | Allergen Sol i 3Translate to AllerML | 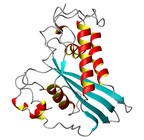 | 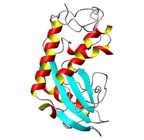 | | Allergen | Sol i 3 | | Type | insects | | Species - Systematic Name | Solenopsis invicta | | Species - Common Name | fire ant | | Keywords | Venom allergen III
| | Class | IUIS |
| 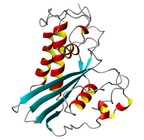 |
| Sol i 3 - PubMed | Reference | | Reference 1 | Hoffman DR, Allergens in Hymenoptera venom XXIV: the amino acid sequences of imported fire ant venom allergens Sol i II, Sol i III, and Sol i IV. J Allergy Clin Immunol. 1993 Jan;91(1 Pt 1):71-8. |
Sol i 3 - Protein Sequences
| Source | Link to Source | View Sequence | FASTA@SDAP | BLAST@NCBI | BLAST@ExPASy | PROSITE@PIR | | SwissProt | P35778 | Go! | Go! | |
|

