
SDAP Home Page
SDAP Overview
SDAP All
SDAP Food
Use SDAP All
Search SDAP All
List SDAP All
SDAP Food
SDAP Tools
FAO/WHO Allergenicity Test
FASTA Search in SDAP
Peptide Match
Peptide Similarity
Peptide-Protein PD Index
Aller_ML, Allergen Markup Language
List SDAP
About SDAP
General Information
Manual
FAQ
Publications
Who Are We
Advisory Board
Allergy Links
Our Software Tools
MPACK
FANTOM
GETAREA
NOAH/DIAMOD
MASIA
PCPMer
InterProSurf
EpiSearch
Protein Databases
PDB
MMDB - Entrez
SWISS-PROT
NCBI - Entrez
PIR
Protein Classification
CATH
CE
FSSP
iProClass
ProtoMap
SCOP
TOPS
VAST
Bioinformatics Servers
@TOME
BLAST @ NCBI
BLAST @ PIR
FASTA @ PIR
Peptide Match @ PIR
ClustalW @ BCM
ClustalW @ EMBL - EBI
ClustalW @ PIR
Bioinformatics Tools
Cn3D
MolMol
Bioinformatics Links
Bioinformatics Links Directory
|
SDAP - All Allergens
Allergen group: Ara h 2 | Allergen Ara h 2.0101Translate to AllerML | 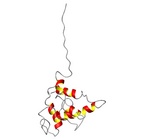 | 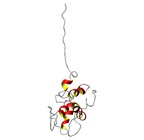 | | Allergen | Ara h 2.0101 | | Type | foods | | Species - Systematic Name | Arachis hypogaea | | Species - Common Name | peanut | | Keywords | Conglutin | | Class | IUIS |
| 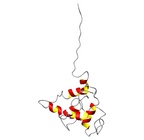 |
| Ara h 2.0101 - PubMed | Reference | | Reference 1 | Kleber-Janke T, Crameri R, Appenzeller U, Schlaak M, Becker WM. Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int Arch Allergy Immunol. 1999 Aug;119(4):265-74. doi: 10.1159/000024203. PMID: 10474031 | | Reference 2 | Chatel JM, Bernard H, Orson FM. Isolation and characterization of two complete Ara h 2 isoforms cDNA. Int Arch Allergy Immunol. 2003 May;131(1):14-8. doi: 10.1159/000070429. PMID: 12759484 | | Reference 3 | Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, Helm RM, West CM, Bannon GA. Identification and mutational analysis of |
|

